This is a brief post because I am no expert, and it is likely I am not the first person to think of using a binary encoding to significantly increase the throughput of viral RNA tests.
Somebody with more experience, please confirm on hackernews (https://news.ycombinator.com/item?id=22612139)
Now that Covid-19 is out, we need to figure out how to detect all infected people so that uninfected people can return to work/school/life. Further, we need continually test everyone who is assumed uninfected weekly to prevent future outbreaks. This test cannot be an antibody test, as an infected person may be contagious before the immune system has created the antibodies. The only current way to detect the virus in an asymptomatic individual is RT-PCR of the viral RNA.
Let's take Denmark, where I now live. That's 5 million people and therefore tests per week. If the current cost of a viral RNA test is $200, that's $1 billion per week or $52 billion per year. These tests are usually done on 96 well plates with 2 control wells. That leaves 94 tests per plate (which can be thought of as batch size for tests).
However, the assumption is that currently most people are not infected. This is especially true if our goal is to weekly test the people returning to daily life who are assumed to be uninfected. Therefore, most of the wells are "wasted" on uninfected individuals.
Here's how to test 4731 people per 96 well plate: take two samples from each individual, and mix the samples in a binary encoding scheme that assign 2 bits of a 94-bit number to each person.
To illustrate, consider first this how to test 6 people using 4 sample wells (forget the controls for now). If each sample well is either red for positive or blue for negative, there are 6 ways to color the wells such that exactly 2 sample wells are positive.
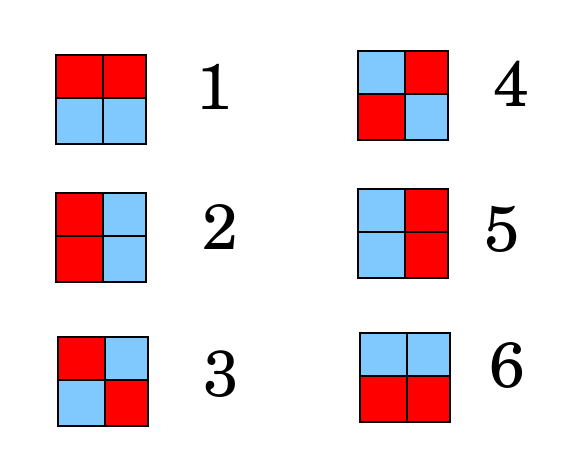
The stategy is to assign each person to one of the above colorings, and then in each well mix the samples from testees such that if one (and only one) person is positive, the resulting coloring will match the assigned person. To do this, place each person's two samples in the wells as shown below.
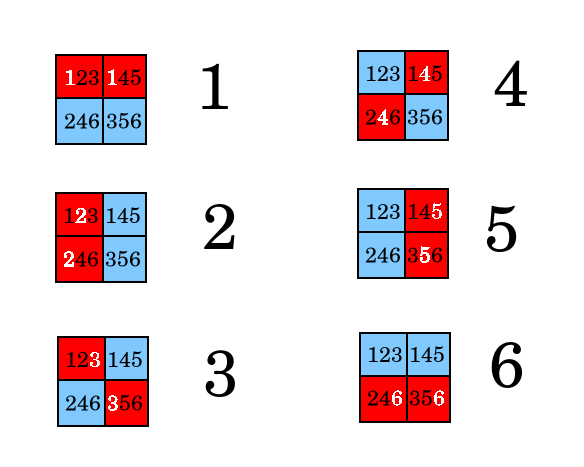
The encoding is equivelent to enumerating the 6 binary numbers between 0 and 15 with two set bits, and then assigning one person to each of those numbers. With 4 wells, we can test 4 choose 2 people = 6. With 94 wells, we can test 94 choose 2 people = 4731.
The PCR step (including preparation) is the bottle-neck in the testing process. For the 94 well method to work, each well would need to combine samples from 93 people, such that 8742 samples are combined to make a single 96 well plate (4731 people times 2 samples each = 8732 samples). The good news is that the test samples can be combined into the 94 groups (one for each well) right at the beginning of the process (literally, each group of 93 swabs could be put together into a large beaker and mixed). All other downstream operations then only need to be done once per well.
We need a mechanical device which can automatically sort the samples into well groups. Correctly combining 4731 samples is not something that can be done by hand. However, compared to the PCR machines and magical reagants, this sorting machine should be relatively simple to build. The reward is a 46.5 times increase in testing throughput.
Yes, there is also the possibility that more than one of the 4731 people is infected, and in this case you would get 3 or more positive wells. Still, this method can be used to determine a subset of samples which need individual retesting. However, if most samples are assumed to be uninfected, this is not a huge issue.